Murphy Lab
HomeInformation
People
Addresses
Cytometry Development Workshop
FCS API
Research
Projects
Publications
Software
Presentations
Flow Cytometry
Data
Download
Affiliations
Carnegie Mellon University
Computational Biology Department
Center for Bioimage Informatics
Biological Sciences Department
Biomedical Engineering Department
Machine Learning Department
MBIC
[an error occurred while processing this directive]
Internal
InformationLab FAQ
Lab Research Topics
Services
TypIX
Web Pages
Web Page
Template
Visitation Statistics
Important Links
Software and Documentation
Matlab Docs
Khoros Docs
TN-Image Docs
Michael Boland's thesis
SImEC - Statistical Imaging Experiment Comparator
Objective evaluation of differences in protein subcellular distribution
Edward J.S. Roques and Robert F. Murphy
Department of Biological Sciences, Biomedical Engineering Program, and Center for Light Microscope Imaging and Biotechnology, Carnegie Mellon University, Pittsburgh PA 15213/USA
Introduction
In order to compare a change in a protein's subcellular localization following a change in expression or in environmental conditions it is first necessary to obtain image sets of the initial and final distribution. Visual analysis of these image sets may prove inadequate for quantitative statements regarding a change in protein localization. SImEC performs a quantitative analysis of the image sets and returns a degree of similarity between the two distributions and whether the protein localization can be regarded as being from the same class, within a statistical level of confidence.
Method
Previous work from our laboratory has described a method for the numerical description of protein localization according to three sets of features: Zernike moment features, Haralick texture features, and features specifically derived for analysis of subcellular location. These features have been shown to be capable of distinguishing the major subcellular organelles and structures of cultured cells. These features can be combined in different ways to form different Subcellular Location Feature (SLF) sets.
Example
HeLa cell images showing the distributions of the Golgi proteins giantin and gpp130 that were collected as part of our previous classification project were used to illustrate the SImEC service. Parallel images showing the distributions of DNA were used, along with hand-specified polygonal regions that isolated an individual cell in each image. Over 80 images for each protein were processed by SImEC using the SLF6 feature set with a confidence level of 0.05. The results generated by SImEC are available here. Examples of the images are shown below.
| Giantin | gpp130 | |
| Protein Image |  |  |
| DNA Image | 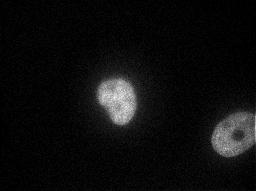 | 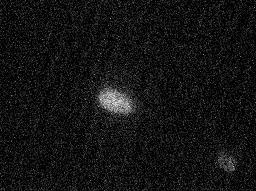 |
| Crop Image |  |  |
Last Updated: 01 Dec 2004
Copyright © 1996-2019 by the Murphy Lab, Computational Biology Department at Carnegie Mellon University