Murphy Lab
HomeInformation
People
Addresses
Cytometry Development Workshop
FCS API
Research
Projects
Publications
Software
Presentations
Flow Cytometry
Data
Download
Affiliations
Carnegie Mellon University
Computational Biology Department
Center for Bioimage Informatics
Biological Sciences Department
Biomedical Engineering Department
Machine Learning Department
MBIC
[an error occurred while processing this directive]
Internal
InformationLab FAQ
Lab Research Topics
Services
TypIX
Web Pages
Web Page
Template
Visitation Statistics
Important Links
Software and Documentation
Matlab Docs
Khoros Docs
TN-Image Docs
Michael Boland's thesis
Murphy Lab - Typical Image Selection
Mia K. Markey, former undergraduate in Biological Sciences
Michael V. Boland, former graduate student in Biomedical Engineering
Introduction
Advances in microscopy (especially fluorescence microscopy) over the past decade have made common the collection of large numbers of digital images as part of biological studies. However, methods for automated analysis of such images have lagged behind hardware developments. One area that has received little attention is the choice of a representative image from a set. Scientists wishing to communicate the essential characteristics of a pattern (such as the immunofluorescence distribution for a newly-discovered protein) currently must make a subjective choice of one or two images to publish. We have therefore developed and characterized methods for choosing a typical image from a set, with an emphasis on images commonly encountered in cell biology.
Approach
Choosing a typical image from a set using the methods we have developed involves three steps: calculating numerical features for each image that are insensitive to translation and rotation of a cell (or other object) within an image and that are believed to summarize useful information about the image defining a measure of similarity between images as a function of the features ranking the images by the distance of each image from the center of the image distribution or by the summed distance to all other images. We have explored two types of features, those based on measures of image texture and those based on Zernike polynomial moments. Different methods for estimating distance between images have also been explored.
Results
To evaluate various methods, we have used a collection of images of Chinese Hamster Ovary cells that was generated as part of our work on classification of protein localization patterns. This collection is available for download. The images depict the patterns of a Golgi complex protein (giantin), a lysosomal membrane protein (LAMP2), a cytoskeletal protein (tubulin) and DNA. As a starting point, we proposed two criteria for evaluating particular methods for calculating typicality based on the use of test data sets comprised of mixtures of images containing more than one type of pattern. The results indicate the importance of using distance measures that are insensitive to the presence of outliers (i.e., atypical images). Good results were obtained using what we have termed HTFR typicality (Haralick Texture Features, Robust estimation of distance). As an illustration of the results, the figure below shows images of the Golgi protein giantin that were selected as most and least typical using one or both of the methods we developed. The images considered most typical (A-D) all show a compact Golgi complex, while those considered least typical tend to be dispersed and punctate (E-H).
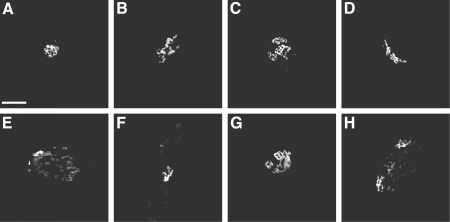
A selection of the most (A-D) and least (E-H) typical giantin images, as determined using various methods (see the reference below for details). Scale bar = 10 microm.
Conclusions
In addition to being used for selecting images for publication, the approach described has potential applications when displaying representative images in Internet-based image databases and during image acquisition by automated microscopes.
Typical Image Chooser (TypIC)
Investigators wishing to use the methods we have developed may upload an image collection and request that the images be ranked by typicality. The collection must include at least 35 images for the HTFR method to work adequately.
Reference
Markey, M.K., Boland, M.V., and Murphy, R.F. (1999) Towards Objective Selection of Representative Microscope Images. Biophysical Journal 76:2230-2237.
Research Credits
This work was supported in part by research grant RPG-95-099-03-MGO from the American Cancer Society (R.F.M.), by Carnegie Mellon's Undergraduate Research Initiative and Howard Hughes Medical Institute Undergraduate Education Program (M.K.M.), by NSF grant BIR-9217091, and by NSF Science and Technology Center grant MCB-8920118. M.V.B. was supported by NIH training grant T32 GM08208 and by NSF training grant BIR-9256343.
Last Updated: 01 Dec 2004
Copyright © 1996-2019 by the Murphy Lab, Computational Biology Department at Carnegie Mellon University