Murphy Lab
HomeInformation
People
Addresses
Cytometry Development Workshop
FCS API
Research
Projects
Publications
Software
Presentations
Flow Cytometry
Data
Download
Affiliations
Carnegie Mellon University
Computational Biology Department
Center for Bioimage Informatics
Biological Sciences Department
Biomedical Engineering Department
Machine Learning Department
MBIC
[an error occurred while processing this directive]
Internal
InformationLab FAQ
Lab Research Topics
Services
TypIX
Web Pages
Web Page
Template
Visitation Statistics
Important Links
Software and Documentation
Matlab Docs
Khoros Docs
TN-Image Docs
Michael Boland's thesis
The Architecture of the SLIF System
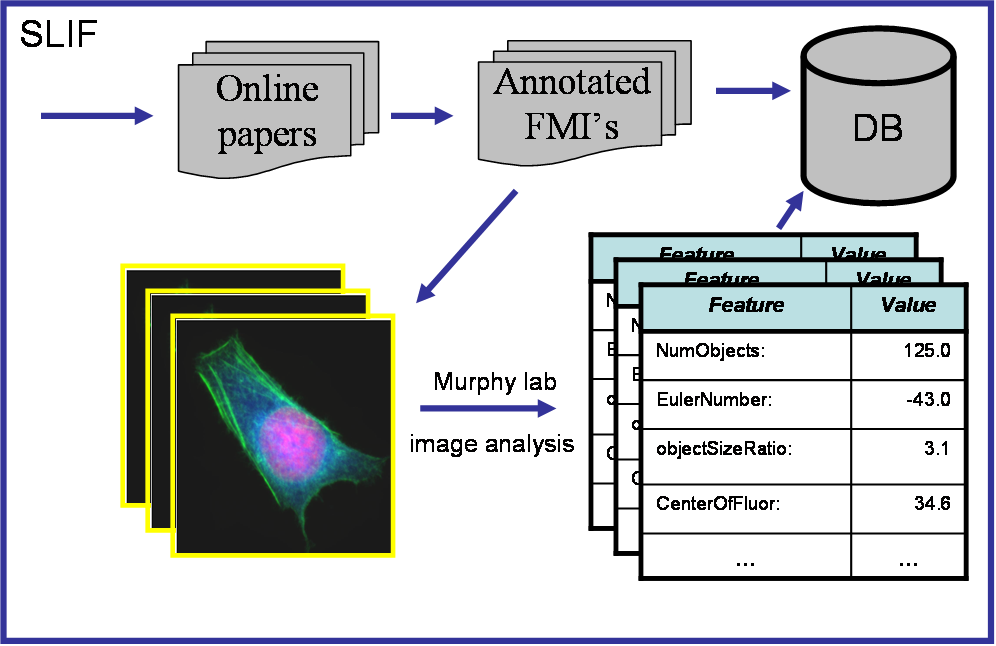
| SLIF takes on-line papers and scans them
for figures that contain fluorescence microscope images (FMIs).
Figures typically contain multiple FMIs, to SLIF must segment
these images into individual FMIs. When the FMI images are extracted,
annotations for the images (for instance, names of proteins and
cell-lines) are also extracted from the accompanying caption
text. Protein annotation are also used to link to external databases,
such as the Gene Ontology DB.
Extracted FMIs are processed to find subcellular location features (SLFs), and the resulting analyzed, annotated figures are stored in a database, which is accessible via SQL queries. |
 |
|
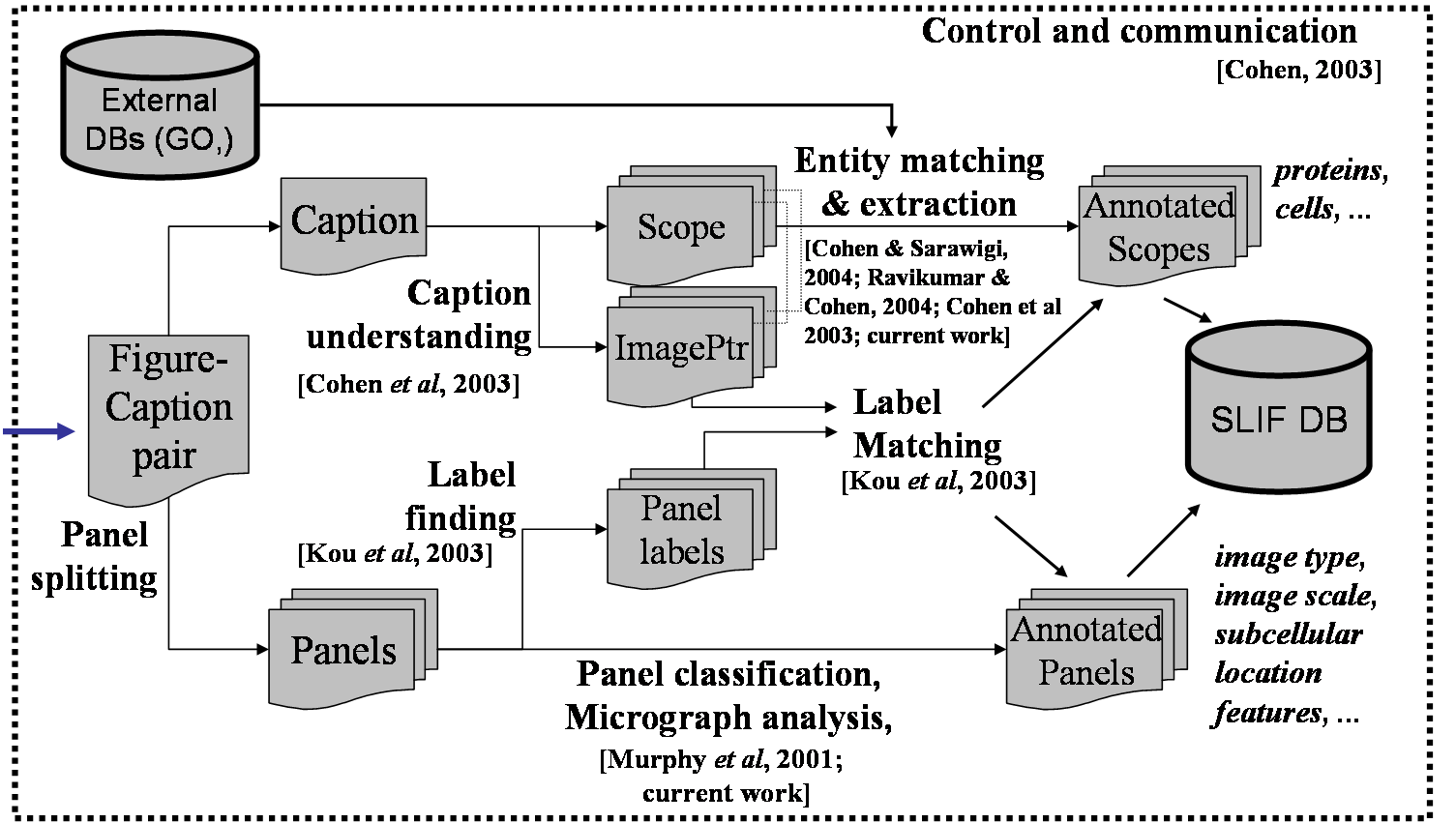
The diagram above shows the SLIF process in outline, and the diagram to the left shows the process in more detail. (Clicking on any diagram will enlarge it.) The more detailed process includes: segmentation of images into "panels"; panel classification, to find FMIs; segmentation of the caption, to find which portions of the caption apply to which panels; text-based entity extraction; matching of extracted entities to database entries; extraction of panel labels from text and figures; and alignment of the text segments to the panels. Publications that discuss these steps in detail are indicated in the detailed diagram. |
  The current
SLIF database is populated with data from publications from The Proceedings of the National Academy of Science
and Biomed Central.
The current
SLIF database is populated with data from publications from The Proceedings of the National Academy of Science
and Biomed Central.
| |